Genomic Breeding
Genomic selection-assisted breeding mode
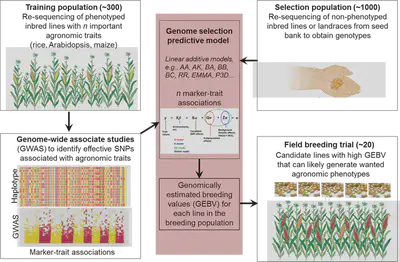
Compared to molecular marker-assisted breeding (MAB), genomic selection (GS) is a new frontier that uses large numbers of SNP markers to predict breeding values to select candidate lines for hybridization. This process involves building a training population with known genotypes and phenotypes to construct GS model and derive the most optimal parameters for the model. Then, for a candidate population, using the genotype as input to predict the trait as output. This method can predict the simulated traits from all possible combination of parental lines in the test population, and then help to select parental lines for actual breeding. We are currently working in rice and maize to improve the GS model, by dissecting effective markers into additive, dominant, over-dominant, and epistatic classes, with quantitative evaluation of marker effects. When training the GS models, we utilize different methods including GBLUP, rrBLUP, Bayesian, and modern machine learning approach such as neural network, deep learning, SVM, RF, decision tree.