Population Genetics
For genetic improvement of crop species, genome-wide association analysis (GWAS) is a rapid way to identify the genes contributing to the variation of important agronomic traits, and the behind principles are population genetics. Since the Wang Lab moved to CAU, we started to work on GWAS of crop germplasm with new methods and new idea. We are working on GWAS in maize, rice, wheat. In a long run, we will construct a database for trait-related genes, tag SNPs, QTLs, and use these sets of information to support crop breeding.
-
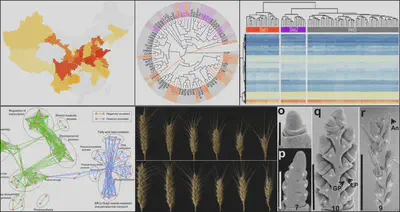
Wheat population
We GWASed a collection of wheat core germplasm with detailed records of spike trait, using ILLUMINA RNA-Seq. With eQTL analysis, we identified a series of candidate genes that may contribute to the wheat spike development. We also provide transgenic evidence to prove the functions of the genes we identified.
-
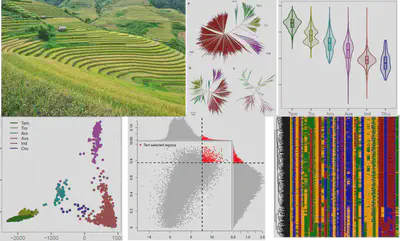
Rice population
Rice, Oryza sativa L., is the staple food for half the world’s population. Based on a resequenced core collection of 3,000 rice accessions from 89 countries, we utilized a series of evolutionary analytical and population methods to determine the frequently selected regions, genes and markers during the domestication and genetic improvement process in rice. From this dataset, our ultimate goal is to construct a database that includes all kinds of trait-related genes, markers and regions to provide a scalable, dynamic environment for rice researcher to extract important genes for breeding purpose.